Investigating Larval Release Triggers in the Native Oyster: University of Glasgow Masters Research

Project Motivation: The Quest to Understand Swarming Timing
In the 1800s, dredging led to a 90% decline in the UK’s native oyster populations. Since larvae need shell materials to settle and there are fewer oyster beds available, they rely on staying close to their birthplaces. Restoration projects add shell materials to create more settling spots, but timing must be perfect—too early, and you risk attracting unwanted species; too late, and you miss peak larval releases, along with the biofilm buildup that aids settlement. Pinpointing swarming could be key to bringing native oysters back from the brink.
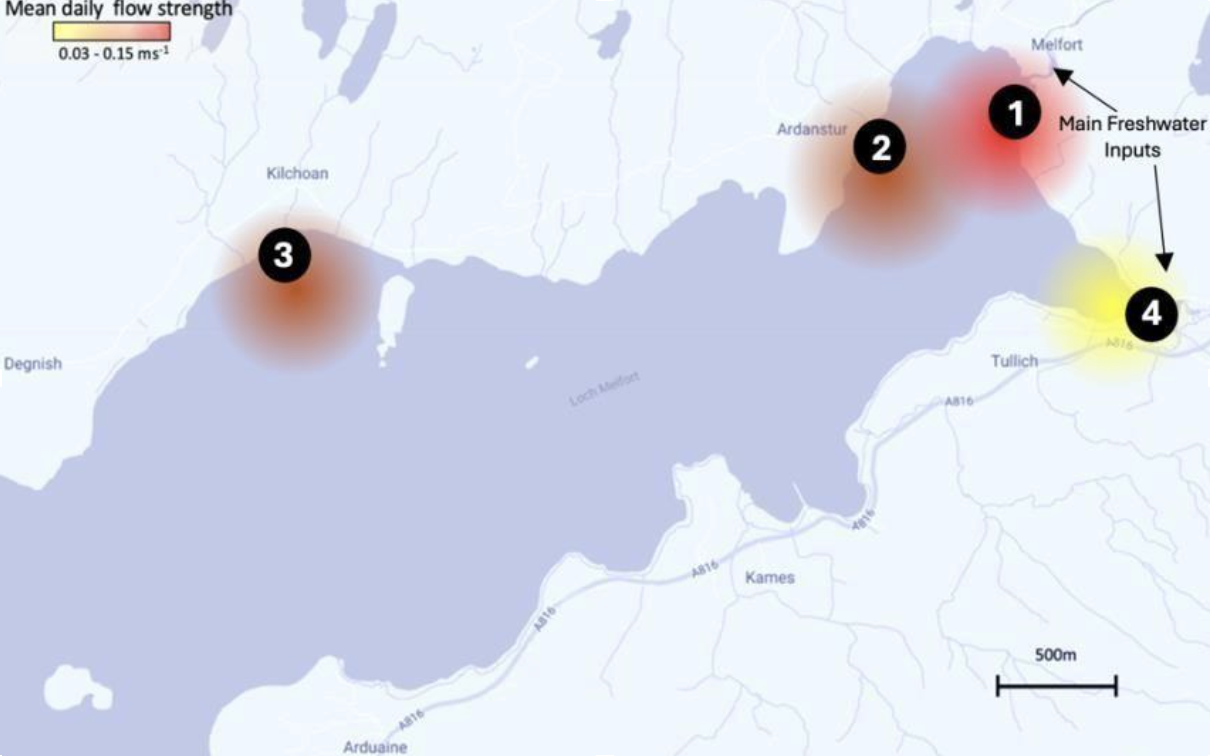
Previous studies on swarming timing are often limited to single study sites and show conflicting views in tidal versus non-tidal flow waters. To broaden our perspective, we studied four sites at Kilchoan Estate with varying freshwater proximity and flow strengths. Because oyster larvae resemble other bivalves, we combined qPCR eDNA analysis with larval counts for precise identification. After
confirming eDNA’s effectiveness in controlled settings, we aimed to test its application in natural environments, further hoping to provide support for larval counts used to identify swarming triggers.
My Research Objectives:
- Identify swarming triggers through larval count monitoring.
- Assess the use of eDNA as a swarming monitoring tool in natural environments.
Data collection:
We collected larval counts, eDNA, and environmental data across four Kilchoan Estate oyster release or presence sites in Loch Melfort. Sampling ran from May to July 2024, carefully timed to coincide with low tides across four tidal phases: spring, spring-to-neap, neap, and neap-to-spring tides.

What We Discovered:
Swarming Triggers?
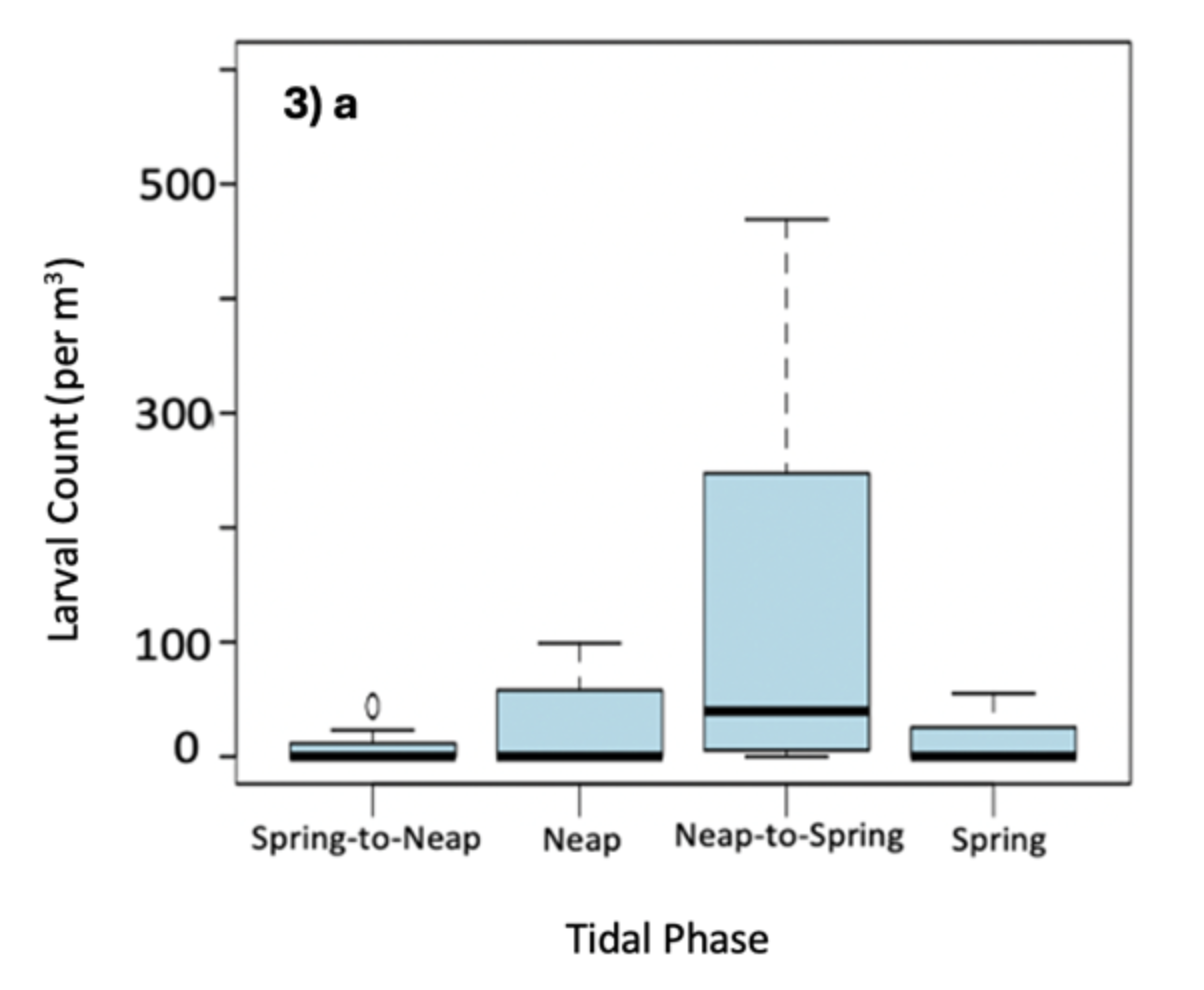
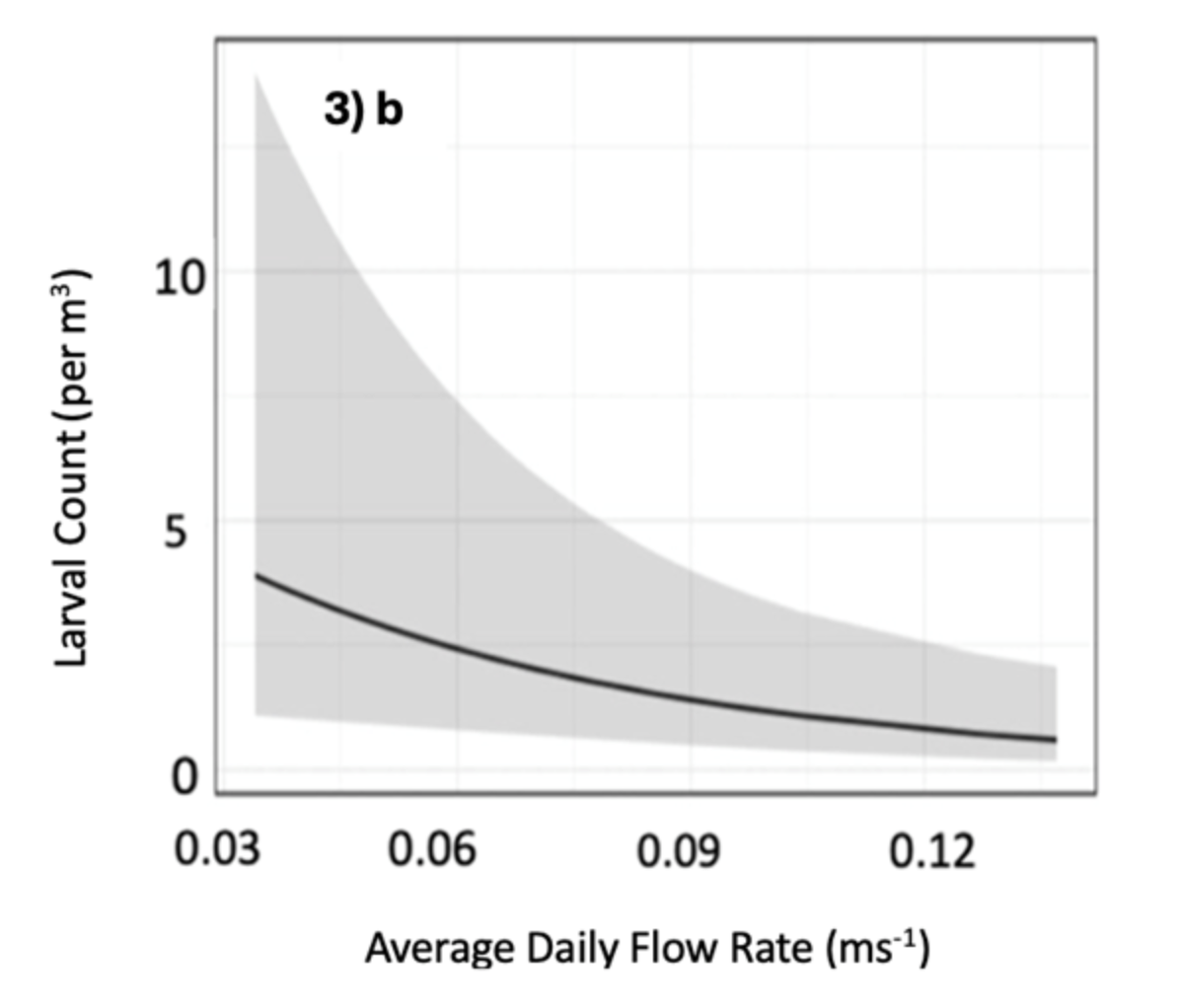
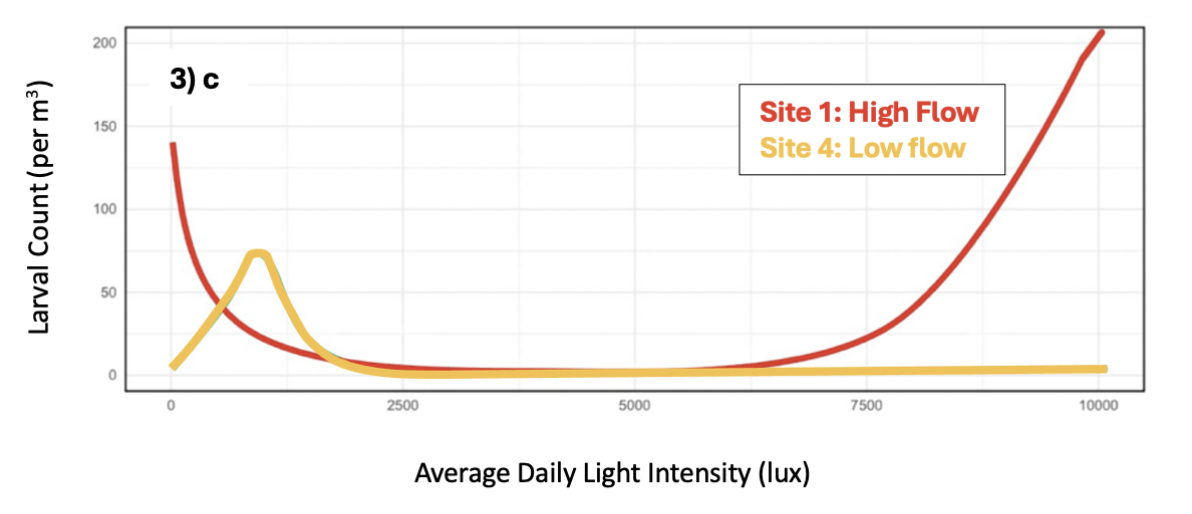
Figure 3. Significant swarming predictors identified through AIC selection. Shows: a) tidal phase (spring-to-neap, neap, neap-to-spring, or spring), b) average daily flow rate (m/s), and c) average daily light intensity (lux) in relation to predicted larval counts/m3 water. Predictions for tidal phase and flow rate are generated across all sites, while light intensity is significant only at sites 1 and 4, colour-coded by flow type.
- Swarming mainly occurs during neap-to-spring tides and low daily flow rates, which likely aid retention and reduce coastal stranding. However, high flow might also dilute detection, making its role as a driver uncertain.
- Daily light intensity influenced larval counts variably by site: low light triggered releases in a low-flow site, while a high-flow site showed bimodal low/high light releases. Since light controls larval vertical migration and dispersal, findings suggest swarming timing relative to light could be optimized differently in flow-challenged sites, though further research is needed.
eDNA – A Promising Tool?
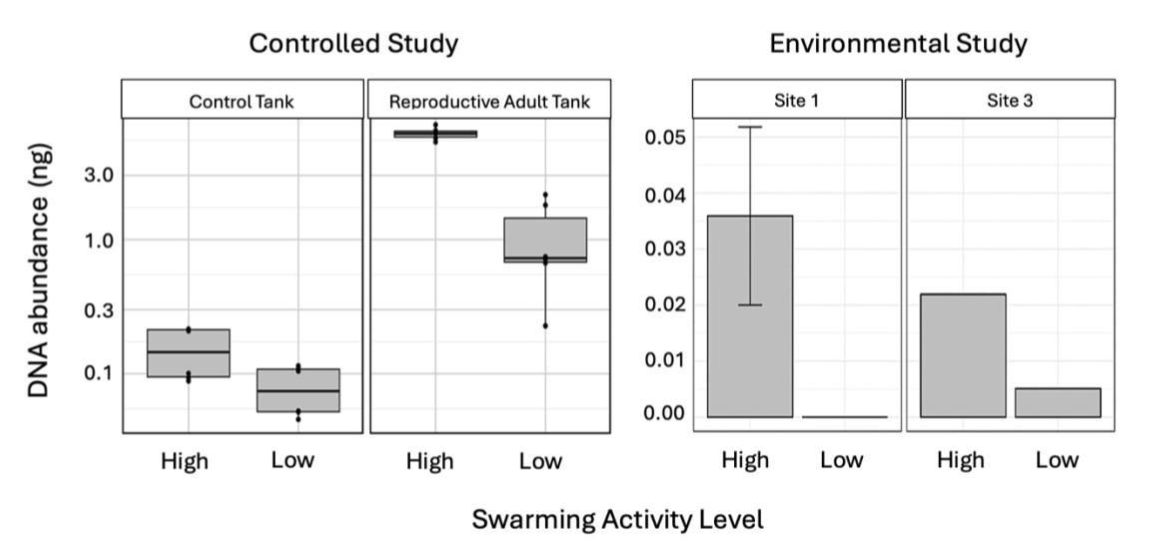
Conclusion and Wider Implications:
This study provides valuable insights for improving oyster restoration by optimizing cultch deployment and larval settlement. By considering environmental triggers such as tidal phases, flow rates, and light intensity, we can better predict larval releases. While eDNA shows promise as a swarming monitoring tool in natural settings, further research is needed to refine its use in dynamic environments.
Final Words and Mentions:
I’ve had a fantastic time at Kilchoan Estate and will always cherish the experience. A huge thank you to Marnik Van Cauter for his exceptional support, and to other members of staff, in particular Laura Dawson and Luke Senior. I’m also grateful to the landowners for allowing my research at the estate – I feel very fortunate. Lastly, a special shoutout to my supervisor, Dr. Anna McGregor, for her support throughout the project, and to Jack Roberts, my data collection buddy, for his help with sampling whilst also working on his own research at the estate.